|
|
| (2 intermediate revisions by the same user not shown) |
| Line 1: |
Line 1: |
| | Note: We do not remove the reads that are suppose to be filtered by the solexa pipeline in version 1.4. However, reads suppose to be filtered are marked using a binary system of 1 = Good/Not Filtered and 0 = Bad/Filtered within the read ID. This information is in the quality score/FASTQ files. | | Note: We do not remove the reads that are suppose to be filtered by the solexa pipeline in version 1.4. However, reads suppose to be filtered are marked using a binary system of 1 = Good/Not Filtered and 0 = Bad/Filtered within the read ID. This information is in the quality score/FASTQ files. |
| | | | |
| | + | == '''''Sequencing Quality Control Based On FASTQ (Basecalls and quality scores)''''' || '''''[[ELANDQC|Go to Sequencing Quality Control Based On ELAND Alignments]]''''' == |
| | | | |
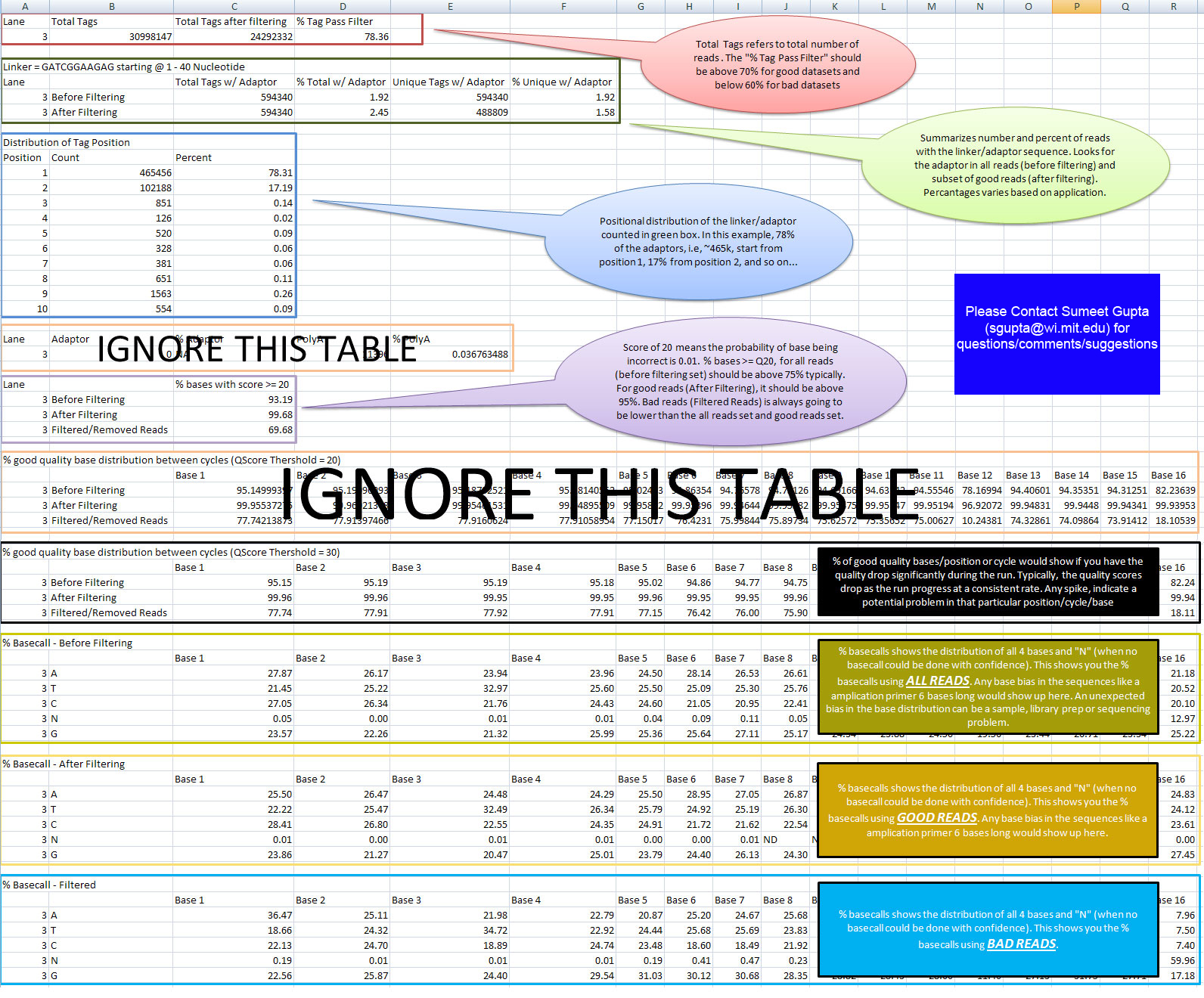
| − | = QCReport Format =
| + | [[Image:FASTQ_QC.jpg]] |
| − | | |
| − | {| class="wikitable" style="font-style:italic; font-size:120%; width:100%; border:2px solid white; height:100px" align="center"
| |
| − | |-
| |
| − | | [[image:QualityReport.jpg|QCReport using Base Quality|center|thumb|500px]]
| |
| − | |
| |
| − | | [[image:ELAND_QC.JPG|ELAND based QC|center|thumb|500px]]
| |
| − | |-
| |
| − | |}
| |
| − | | |
| − | | |
| − | == Yellow Box (QCReport using Base Quality) ==
| |
| − | Column 1: Lane #/Sample id <br>
| |
| − | Column 2: Total # of unique reads (i.e. if a read is repeated in the dataset, it is not counted)<br>
| |
| − | Column 3: Total # of unique reads AFTER FILTERING (Please refer to [http://jura.wi.mit.edu/genomecorewiki/index.php/Sumeet_Gupta#Do_we_filter_.22bad.22_reads_from_the_final_dataset.3F_.5B06.2F19.2F09.5D| FAQ] for questions on filtering)<br>
| |
| − | Column 4: Total # of reads in the dataset<br>
| |
| − | Column 5: Total # of reads IN FILTERED READS (Please refer to [http://jura.wi.mit.edu/genomecorewiki/index.php/Sumeet_Gupta#Do_we_filter_.22bad.22_reads_from_the_final_dataset.3F_.5B06.2F19.2F09.5D| FAQ] for questions on filtering)<br>
| |
| − | | |
| − | == Brown Box (QCReport using Base Quality) ==
| |
| − | Column 1: Lane #/Sample id<br>
| |
| − | Column 2: Type of Dataset (filtered or not) (Please refer to the Solexa Sample Processing Details OR FAQ for questions on filtering)<br>
| |
| − | Column 3: Total # of reads in the dataset with Tag/Linker <br>
| |
| − | Column 4: PERCENT Total # of reads in the dataset with Tag/Linker <br>
| |
| − | Column 5: Unique # of reads in the dataset with Tag/Linker (Please refer to the FAQ for questions on filtering)<br>
| |
| − | Column 5: PERCENT Unique # of reads in the dataset with Tag/Linker (Please refer to the FAQ for questions on filtering)<br>
| |
| − | | |
| − | == Green Box (QCReport using Base Quality) ==
| |
| − | Column 1: Position on the Reads<br>
| |
| − | Column 2: Total # of Adaptor/Linker/ Reads Starting at Position specified in column 1<br>
| |
| − | Column 3: PERCENT Total # of Adaptor/Linker/ Reads Starting at Position specified in column 1<br>
| |
| − | | |
| − | == Blue Box (QCReport using Base Quality) ==
| |
| − | Column 1: Lane #/Sample id<br>
| |
| − | Column 2: Total # of Adaptor Reads<br>
| |
| − | Column 3: PERCENT Total # of Adaptor Reads<br>
| |
| − | Column 4: Total # of PolyA Reads<br>
| |
| − | Column 5: PERCENT Total # of PolyA Reads<br>
| |
| − | | |
| − | == Grey Box (QCReport using Base Quality) ==
| |
| − | Column 1: Lane #/Sample id<br>
| |
| − | Column 2: Type of Dataset (filtered or not) (Please refer to the FAQ for questions on filtering)<br>
| |
| − | Column 3: Percentage of bases with a quality score of atleast 20 (i.e. the probability of base call being incorrect is 1 in a 100)<br>
| |
| − | | |
| − | == Purple Box (QCReport using Base Quality) ==
| |
| − | Column 1: Lane #/Sample id<br>
| |
| − | Column 2: Type of Dataset (filtered or not) (Please refer to the FAQ for questions on filtering)<br>
| |
| − | Column 3 and further: Percentage of bases with a quality score of atleast 20 in that cycle/position.<br>
| |
| − | | |
| − | == Yellow Box (ELAND based QC) ==
| |
| − | Column 1: Files
| |
| − | Column 2: Genome Used
| |
| − | Column 3: Total Reads
| |
| − | Column 4: Reads Kept (Column 3 - Column 5)
| |
| − | Column 5: Solexa Linker(Reads Removed)
| |
| − | Column 6: % Removed
| |
| − | Column 7: # of Reads that align Unique
| |
| − | Column 8: % of Reads that align Unique
| |
| − | Column 9: # of Reads fail to align because of too many N's
| |
| − | Column 10: % reads w/ many N's
| |
| − | Column 11: Reads with Multiple Matches
| |
| − | Column 12: % reads w/ multi-match
| |
| − | Column 13: Reads with No Match
| |
| − | Column 14: % reads w/ no-match
| |
| − | | |
| − | == Green Box (ELAND based QC) ==
| |
| − | Break down of the unique reads in U0, U1, U2.... and so on.
| |
| − | | |
| − | == Blue Box (ELAND based QC) ==
| |
| − | PERCENT Break down of the unique reads in U0, U1, U2.... and so on.
| |
| − | | |
| − | == Brown Box (ELAND based QC) ==
| |
| − | Number of mismatches at each position i.e. for a 36 base run, number of mismatches for position 1, position 2 ... and so on to position 36.
| |
| − | | |
| − | == Grey Box (ELAND based QC) ==
| |
| − | PERCENT mismatches at each position i.e. for a 36 base run, PERCENT mismatches for position 1, position 2 ... and so on to position 36.
| |
Note: We do not remove the reads that are suppose to be filtered by the solexa pipeline in version 1.4. However, reads suppose to be filtered are marked using a binary system of 1 = Good/Not Filtered and 0 = Bad/Filtered within the read ID. This information is in the quality score/FASTQ files.