Difference between revisions of "QCOutputFormat"
| (4 intermediate revisions by the same user not shown) | |||
| Line 1: | Line 1: | ||
| + | Note: We do not remove the reads that are suppose to be filtered by the solexa pipeline in version 1.4. However, reads suppose to be filtered are marked using a binary system of 1 = Good/Not Filtered and 0 = Bad/Filtered within the read ID. This information is in the quality score/FASTQ files. | ||
| + | |||
| + | |||
= QCReport Format = | = QCReport Format = | ||
{| class="wikitable" style="font-style:italic; font-size:120%; width:100%; border:2px solid white; height:100px" align="center" | {| class="wikitable" style="font-style:italic; font-size:120%; width:100%; border:2px solid white; height:100px" align="center" | ||
|- | |- | ||
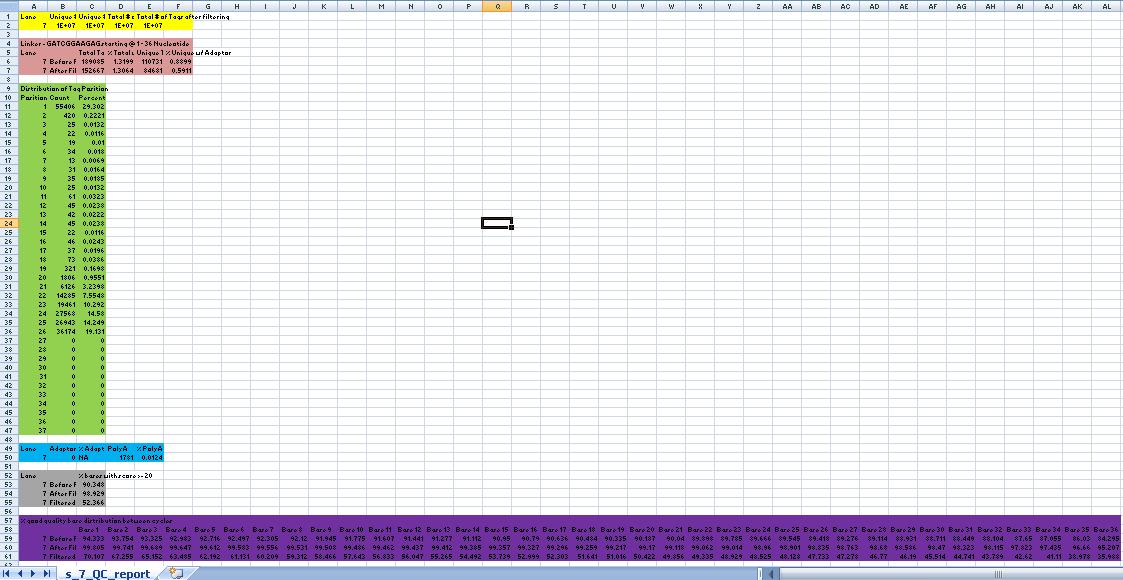
| − | | [[image:QualityReport.jpg|center|thumb|500px]] | + | | [[image:QualityReport.jpg|QCReport using Base Quality|center|thumb|500px]] |
| + | | | ||
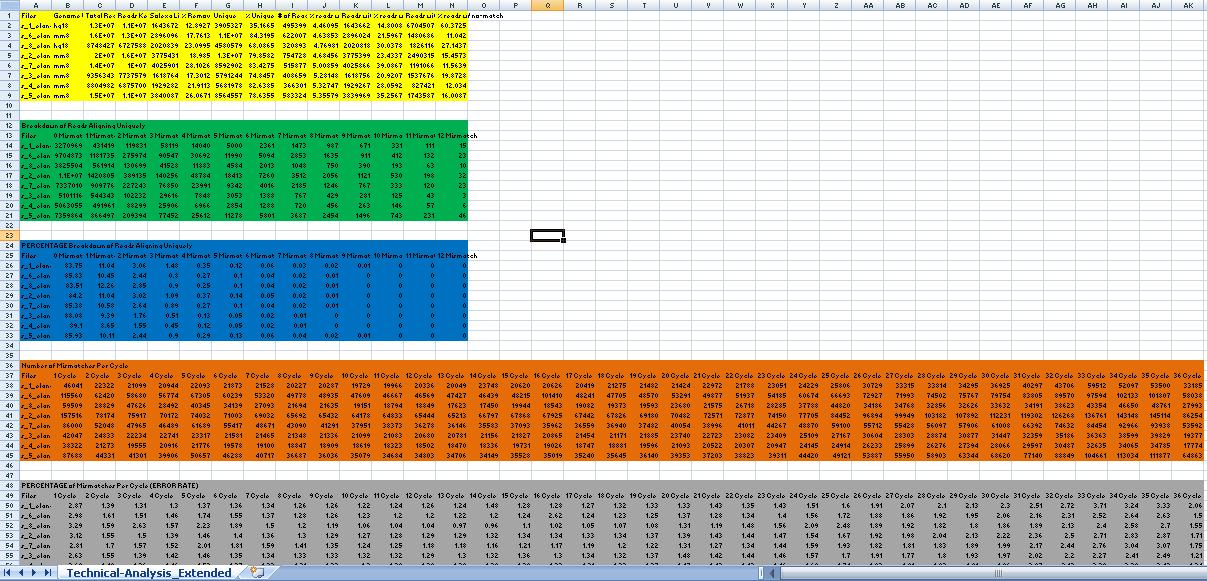
| + | | [[image:ELAND_QC.JPG|ELAND based QC|center|thumb|500px]] | ||
|- | |- | ||
|} | |} | ||
| − | == Yellow Box == | + | == Yellow Box (QCReport using Base Quality) == |
Column 1: Lane #/Sample id <br> | Column 1: Lane #/Sample id <br> | ||
Column 2: Total # of unique reads (i.e. if a read is repeated in the dataset, it is not counted)<br> | Column 2: Total # of unique reads (i.e. if a read is repeated in the dataset, it is not counted)<br> | ||
| − | Column 3: Total # of unique reads AFTER FILTERING (Please refer to | + | Column 3: Total # of unique reads AFTER FILTERING (Please refer to [http://jura.wi.mit.edu/genomecorewiki/index.php/Sumeet_Gupta#Do_we_filter_.22bad.22_reads_from_the_final_dataset.3F_.5B06.2F19.2F09.5D| FAQ] for questions on filtering)<br> |
Column 4: Total # of reads in the dataset<br> | Column 4: Total # of reads in the dataset<br> | ||
| − | Column 5: Total # of reads IN FILTERED READS (Please refer to | + | Column 5: Total # of reads IN FILTERED READS (Please refer to [http://jura.wi.mit.edu/genomecorewiki/index.php/Sumeet_Gupta#Do_we_filter_.22bad.22_reads_from_the_final_dataset.3F_.5B06.2F19.2F09.5D| FAQ] for questions on filtering)<br> |
| − | == | + | == Brown Box (QCReport using Base Quality) == |
Column 1: Lane #/Sample id<br> | Column 1: Lane #/Sample id<br> | ||
Column 2: Type of Dataset (filtered or not) (Please refer to the Solexa Sample Processing Details OR FAQ for questions on filtering)<br> | Column 2: Type of Dataset (filtered or not) (Please refer to the Solexa Sample Processing Details OR FAQ for questions on filtering)<br> | ||
Column 3: Total # of reads in the dataset with Tag/Linker <br> | Column 3: Total # of reads in the dataset with Tag/Linker <br> | ||
Column 4: PERCENT Total # of reads in the dataset with Tag/Linker <br> | Column 4: PERCENT Total # of reads in the dataset with Tag/Linker <br> | ||
| − | Column 5: Unique # of reads in the dataset with Tag/Linker (Please refer to the | + | Column 5: Unique # of reads in the dataset with Tag/Linker (Please refer to the FAQ for questions on filtering)<br> |
| − | Column 5: PERCENT Unique # of reads in the dataset with Tag/Linker (Please refer to the | + | Column 5: PERCENT Unique # of reads in the dataset with Tag/Linker (Please refer to the FAQ for questions on filtering)<br> |
| − | == | + | == Green Box (QCReport using Base Quality) == |
| + | Column 1: Position on the Reads<br> | ||
| + | Column 2: Total # of Adaptor/Linker/ Reads Starting at Position specified in column 1<br> | ||
| + | Column 3: PERCENT Total # of Adaptor/Linker/ Reads Starting at Position specified in column 1<br> | ||
| + | |||
| + | == Blue Box (QCReport using Base Quality) == | ||
Column 1: Lane #/Sample id<br> | Column 1: Lane #/Sample id<br> | ||
Column 2: Total # of Adaptor Reads<br> | Column 2: Total # of Adaptor Reads<br> | ||
| Line 30: | Line 40: | ||
Column 5: PERCENT Total # of PolyA Reads<br> | Column 5: PERCENT Total # of PolyA Reads<br> | ||
| − | == | + | == Grey Box (QCReport using Base Quality) == |
Column 1: Lane #/Sample id<br> | Column 1: Lane #/Sample id<br> | ||
| − | Column 2: Type of Dataset (filtered or not) (Please refer to the | + | Column 2: Type of Dataset (filtered or not) (Please refer to the FAQ for questions on filtering)<br> |
Column 3: Percentage of bases with a quality score of atleast 20 (i.e. the probability of base call being incorrect is 1 in a 100)<br> | Column 3: Percentage of bases with a quality score of atleast 20 (i.e. the probability of base call being incorrect is 1 in a 100)<br> | ||
| − | == Purple Box == | + | == Purple Box (QCReport using Base Quality) == |
Column 1: Lane #/Sample id<br> | Column 1: Lane #/Sample id<br> | ||
| − | Column 2: Type of Dataset (filtered or not) (Please refer to the | + | Column 2: Type of Dataset (filtered or not) (Please refer to the FAQ for questions on filtering)<br> |
Column 3 and further: Percentage of bases with a quality score of atleast 20 in that cycle/position.<br> | Column 3 and further: Percentage of bases with a quality score of atleast 20 in that cycle/position.<br> | ||
| + | |||
| + | == Yellow Box (ELAND based QC) == | ||
| + | Column 1: Files | ||
| + | Column 2: Genome Used | ||
| + | Column 3: Total Reads | ||
| + | Column 4: Reads Kept (Column 3 - Column 5) | ||
| + | Column 5: Solexa Linker(Reads Removed) | ||
| + | Column 6: % Removed | ||
| + | Column 7: # of Reads that align Unique | ||
| + | Column 8: % of Reads that align Unique | ||
| + | Column 9: # of Reads fail to align because of too many N's | ||
| + | Column 10: % reads w/ many N's | ||
| + | Column 11: Reads with Multiple Matches | ||
| + | Column 12: % reads w/ multi-match | ||
| + | Column 13: Reads with No Match | ||
| + | Column 14: % reads w/ no-match | ||
| + | |||
| + | == Green Box (ELAND based QC) == | ||
| + | Break down of the unique reads in U0, U1, U2.... and so on. | ||
| + | |||
| + | == Blue Box (ELAND based QC) == | ||
| + | PERCENT Break down of the unique reads in U0, U1, U2.... and so on. | ||
| + | |||
| + | == Brown Box (ELAND based QC) == | ||
| + | Number of mismatches at each position i.e. for a 36 base run, number of mismatches for position 1, position 2 ... and so on to position 36. | ||
| + | |||
| + | == Grey Box (ELAND based QC) == | ||
| + | PERCENT mismatches at each position i.e. for a 36 base run, PERCENT mismatches for position 1, position 2 ... and so on to position 36. | ||
Latest revision as of 14:44, 6 July 2010
Note: We do not remove the reads that are suppose to be filtered by the solexa pipeline in version 1.4. However, reads suppose to be filtered are marked using a binary system of 1 = Good/Not Filtered and 0 = Bad/Filtered within the read ID. This information is in the quality score/FASTQ files.
Contents
- 1 QCReport Format
- 1.1 Yellow Box (QCReport using Base Quality)
- 1.2 Brown Box (QCReport using Base Quality)
- 1.3 Green Box (QCReport using Base Quality)
- 1.4 Blue Box (QCReport using Base Quality)
- 1.5 Grey Box (QCReport using Base Quality)
- 1.6 Purple Box (QCReport using Base Quality)
- 1.7 Yellow Box (ELAND based QC)
- 1.8 Green Box (ELAND based QC)
- 1.9 Blue Box (ELAND based QC)
- 1.10 Brown Box (ELAND based QC)
- 1.11 Grey Box (ELAND based QC)
QCReport Format
Yellow Box (QCReport using Base Quality)
Column 1: Lane #/Sample id
Column 2: Total # of unique reads (i.e. if a read is repeated in the dataset, it is not counted)
Column 3: Total # of unique reads AFTER FILTERING (Please refer to FAQ for questions on filtering)
Column 4: Total # of reads in the dataset
Column 5: Total # of reads IN FILTERED READS (Please refer to FAQ for questions on filtering)
Brown Box (QCReport using Base Quality)
Column 1: Lane #/Sample id
Column 2: Type of Dataset (filtered or not) (Please refer to the Solexa Sample Processing Details OR FAQ for questions on filtering)
Column 3: Total # of reads in the dataset with Tag/Linker
Column 4: PERCENT Total # of reads in the dataset with Tag/Linker
Column 5: Unique # of reads in the dataset with Tag/Linker (Please refer to the FAQ for questions on filtering)
Column 5: PERCENT Unique # of reads in the dataset with Tag/Linker (Please refer to the FAQ for questions on filtering)
Green Box (QCReport using Base Quality)
Column 1: Position on the Reads
Column 2: Total # of Adaptor/Linker/ Reads Starting at Position specified in column 1
Column 3: PERCENT Total # of Adaptor/Linker/ Reads Starting at Position specified in column 1
Blue Box (QCReport using Base Quality)
Column 1: Lane #/Sample id
Column 2: Total # of Adaptor Reads
Column 3: PERCENT Total # of Adaptor Reads
Column 4: Total # of PolyA Reads
Column 5: PERCENT Total # of PolyA Reads
Grey Box (QCReport using Base Quality)
Column 1: Lane #/Sample id
Column 2: Type of Dataset (filtered or not) (Please refer to the FAQ for questions on filtering)
Column 3: Percentage of bases with a quality score of atleast 20 (i.e. the probability of base call being incorrect is 1 in a 100)
Purple Box (QCReport using Base Quality)
Column 1: Lane #/Sample id
Column 2: Type of Dataset (filtered or not) (Please refer to the FAQ for questions on filtering)
Column 3 and further: Percentage of bases with a quality score of atleast 20 in that cycle/position.
Yellow Box (ELAND based QC)
Column 1: Files Column 2: Genome Used Column 3: Total Reads Column 4: Reads Kept (Column 3 - Column 5) Column 5: Solexa Linker(Reads Removed) Column 6: % Removed Column 7: # of Reads that align Unique Column 8: % of Reads that align Unique Column 9: # of Reads fail to align because of too many N's Column 10: % reads w/ many N's Column 11: Reads with Multiple Matches Column 12: % reads w/ multi-match Column 13: Reads with No Match Column 14: % reads w/ no-match
Green Box (ELAND based QC)
Break down of the unique reads in U0, U1, U2.... and so on.
Blue Box (ELAND based QC)
PERCENT Break down of the unique reads in U0, U1, U2.... and so on.
Brown Box (ELAND based QC)
Number of mismatches at each position i.e. for a 36 base run, number of mismatches for position 1, position 2 ... and so on to position 36.
Grey Box (ELAND based QC)
PERCENT mismatches at each position i.e. for a 36 base run, PERCENT mismatches for position 1, position 2 ... and so on to position 36.