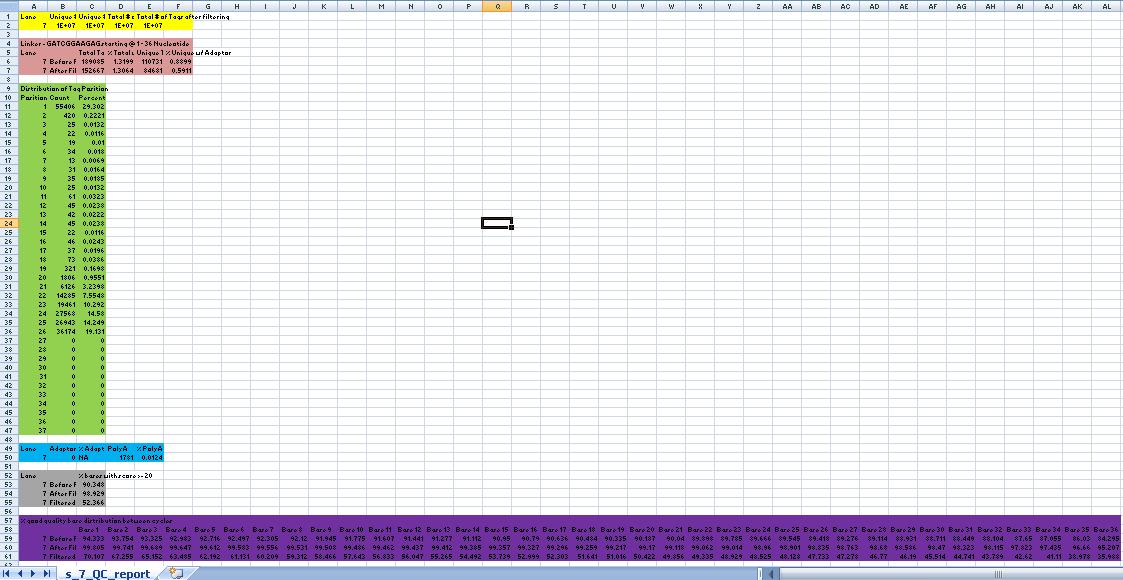
QCOutputFormat
Contents
QCReport Format
Yellow Box
Column 1: Lane #/Sample id
Column 2: Total # of unique reads (i.e. if a read is repeated in the dataset, it is not counted)
Column 3: Total # of unique reads AFTER FILTERING (Please refer to the Solexa Sample Processing Details OR FAQ for questions on filtering)
Column 4: Total # of reads in the dataset
Column 5: Total # of reads IN FILTERED READS (Please refer to the Solexa Sample Processing Details OR FAQ for questions on filtering)
Green Box
Column 1: Lane #/Sample id
Column 2: Type of Dataset (filtered or not) (Please refer to the Solexa Sample Processing Details OR FAQ for questions on filtering)
Column 3: Total # of reads in the dataset with Tag/Linker
Column 4: PERCENT Total # of reads in the dataset with Tag/Linker
Column 5: Unique # of reads in the dataset with Tag/Linker (Please refer to the Solexa Sample Processing Details OR FAQ for questions on filtering)
Column 5: PERCENT Unique # of reads in the dataset with Tag/Linker (Please refer to the Solexa Sample Processing Details OR FAQ for questions on filtering)
Orange Box
Column 1: Lane #/Sample id
Column 2: Total # of Adaptor Reads
Column 3: PERCENT Total # of Adaptor Reads
Column 4: Total # of PolyA Reads
Column 5: PERCENT Total # of PolyA Reads
Purple Box
Column 1: Lane #/Sample id
Column 2: Type of Dataset (filtered or not) (Please refer to the Solexa Sample Processing Details OR FAQ for questions on filtering)
Column 3: Percentage of bases with a quality score of atleast 20 (i.e. the probability of base call being incorrect is 1 in a 100)
Purple Box
Column 1: Lane #/Sample id
Column 2: Type of Dataset (filtered or not) (Please refer to the Solexa Sample Processing Details OR FAQ for questions on filtering)
Column 3 and further: Percentage of bases with a quality score of atleast 20 in that cycle/position.