Difference between revisions of "SequencingFormats"
| Line 1: | Line 1: | ||
We typically provide the following files/formats as part of a typical data package for HiSeq as well as Genome Analyzer Sequencing Data. | We typically provide the following files/formats as part of a typical data package for HiSeq as well as Genome Analyzer Sequencing Data. | ||
| + | |||
| + | <b>WE SPIKE PHIX IN EVERY LANE AND ASSUME THE DE-MULTIPLEXING PROCESS REMOVES IT FROM THE ACTUAL LIBRARY DATA.</b> | ||
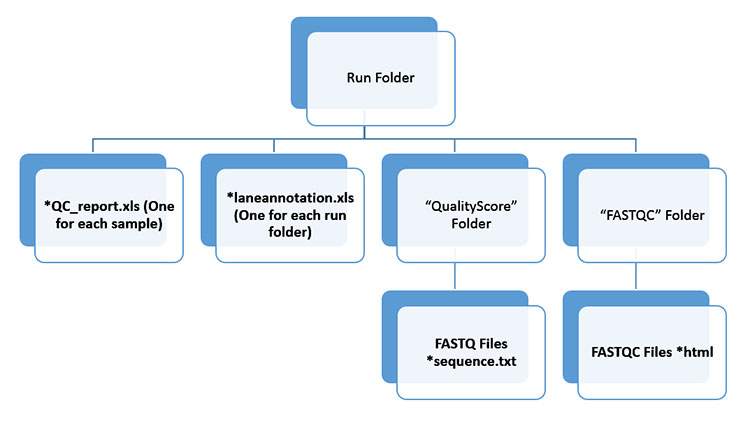
The directory structure for dataset handed over: | The directory structure for dataset handed over: | ||
Revision as of 15:07, 12 January 2017
We typically provide the following files/formats as part of a typical data package for HiSeq as well as Genome Analyzer Sequencing Data.
WE SPIKE PHIX IN EVERY LANE AND ASSUME THE DE-MULTIPLEXING PROCESS REMOVES IT FROM THE ACTUAL LIBRARY DATA.
The directory structure for dataset handed over:
Contents
FASTQ Files (Quality Score Files)
This file format is used frequently at the Sanger Institute to bundle a sequence and its quality data. You will find these files in the "QualityScore" folder of the dataset.
A Illumina FASTQ file normally uses four lines per sequence:
Line 1 - begins with a '@' character and is followed by a sequence identifier
Line 2 - the sequence
Line 3 - begins with a '+' character and is optionally followed by the same sequence identifier
Line 4 - the quality values for each base in the sequence in Line 2
An Example:
@WICMT-SOLEXA_0043:7:1:1500:1199#0/1;1
CTCTGCTGTCTCTTGTGTAAGAAGANANNNCNTTCT
+WICMT-SOLEXA_0043:7:1:1500:1199#0/1;1
fffffffRcfad[ddffa]faaaaBBBBBBBBBBBB
The sequence identifier in the above example is "WICMT-SOLEXA_0043:7:1:1500:1199#0/1;1". Please note that the last 2 characters i.e. ";1" may only be found in datasets sequenced by us because we do not remove the filtered reads but indicate them with a 1 (not filtered i.e. good) or 0 (filtered i.e. bad). Please refer to the FAQ's for information on filtering criteria and what it means.
| WICMT-SOLEXA | Instrument name |
|---|---|
| 0043 | A unique random string for the whole run (pretty much meaningless) |
| 7 | flowcell lane |
| 1 | tile number within the flowcell lane |
| 1500 | 'x'-coordinate of the cluster within the tile |
| 1199 | 'y'-coordinate of the cluster within the tile |
| #0 | index number for a multiplexed sample (0 for no indexing) |
| /1 | the member of a pair, /1 or /2 (paired-end or mate-pair reads only) |
| ;1 | Read was not filtered This bit of information is typically not present in FASTQ files created by Illumina pipeline |
Converting *.tar.gz files to *gz files
We starting creating *.tar.gz files to save the full path (which included the run folder name) with each file. However, later on third party tools like FASTQC don't accept this format. Many of these tools still accept *.gz format. The following single line command can be used to convert *.tar.gz to *gz files:
tar -xOzf xyz.tar.gz | gzip -f > xyz.fq.gz
QC_Report Files
This file summarizes the quality scores (overall as well as per cycle), number of reads and how many were filtered, number of sequences with adaptors and at what position/cycle the adaptor starts as well as the base composition for the entire lane on a cycle by cycles bases for the filtered and unfiltered sets.
Please refer to the QC section for details.
FASTQC Reports
Each FASTQ file goes through our pipeline that generates FASTQC reports. These are handed over with the dataset.
Please refer to the FASTQC Website for details.
FASTQ SCREEN Reports
Each FASTQ file goes through our pipeline that generates FASTQ SCREEN reports. These will be handed over with the datasets starting 1st July 2016.
Please refer to the FASTQ SCREEN Website for details.
laneannotation.xls
This excel has information about which sample/gtc id is present in which lane and what is the barcode assigned to it. A list of columns and description in the file is available below (in the order they are in the excel):
| Column Name | Description |
|---|---|
| Position | Lane on the flowcell |
| SampleType | Type of Sample - Generally the same as selected during sample submission |
| Organism | Generally the same as selected during sample submission |
| GelPrepDate | Ignore, this column is no longer tracked but has not been deleted from the file to keep format consistent |
| SampleName | Generally the same as selected during sample submission |
| Barcode | Barcode associated with the library |
| InsertSizeRange | Size selection range |
| AverageFragmentSize | Avg. Fragment Size based on smear analysis |
| ID | GTC ID issued at submission |
| UserName | The same as selected during sample submission |
| Group | Lab name: The same as selected during sample submission |
| Adaptor | Type of Adaptor used for prep |
| SeqPrimer | Type of Primer used for sequencing i.e. standard or custom |
| SeqType | Read Length |
| ReferenceGenome | Generally the same as selected during sample submission |
| concbyNanoDrop(ng/ul)(orig.) | Ignore, this column is no longer tracked but has not been deleted from the file to keep format consistent |
| concbyqPCR(nM)(DF) | Concentration of the final library stock using Qubit |
| concbyBioA(nM)(DF) | Concentration of the final library stock using BioA/FA |
| concbyQubit(nM)(DF) | Concentration of the final library stock using Qpcr |
| ConcUsed | Concentration used for loading the lane |
| NANODROP BASED DF | Ignore, this column is no longer tracked but has not been deleted from the file to keep format consistent |
| DF | Ignore, this column is no longer tracked but has not been deleted from the file to keep format consistent |
| ulindencktl | Ignore, this column is no longer tracked but has not been deleted from the file to keep format consistent |
| ulloaded | Ignore, this column is no longer tracked but has not been deleted from the file to keep format consistent |
| pMloaded | Amount loaded |
| Equimolar? | Generally the same as selected during sample submission |
| FlowCell | Flow cell ID |
| Sequencer | Sequencer name |
| QC | Passed or Marginal or Failed QC |
| Total Reads | Total reads for the sample |
| % Q30 | Percent base above Q30 |
| % Pass Filer | Percent reads pass filter |
| % Adapter | Percent adaptor |
| % Aligned | Percent aligned |
| % Complexity | Percent Complexity |
| NumberOfPeaks | Number of peaks identified using MACS |
| %RiP | Percent reads in peaks |
| Prep - Input NanoDrop Conc. | Ignore, this column is no longer tracked but has not been deleted from the file to keep format consistent |
| Prep - Input QBIT Conc. | Qubit Conc. In ng/ul |
| Prep - Conc. Used | Sample Conc. Used - Qubit or Nanodrop |
| Prep - Input Amount Used | Total amount of input/sample used for the prep (ng) |
| Prep Method | Prep Method used |
| Prep - AO Input | Adaptor Oligo dilution/Concentration used |
| Prep - PCR Cycles | Number of PCR cycles for Prep |
| Prep QC | Passed or Marginal or Failed Prep QC |
Discontinued File Formats
Tag Count Files
These files are created only on request. They have 4 columns:
Column 1 - Sequence
Column 2 - Number of occurrences of the sequence in column 1 BEFORE filtering
Column 3 - Does the sequence have the 3' adaptor [1 = yes, 0 = No]
Column 4 - Number of occurrences of the sequence in column 1 AFTER filtering
Technical-Analysis Files
This file is a summary of the alignment results. Please refer to the QC section for more information about this file.
You will only receive this file if a genome alignment was requested.
ELAND Alignment Files
These files are created if alignments are requested. They can be found in the "Eland_Results_Extended" folder.
The current file naming convention is ss_<LANE>_eland_extended_<GENOME>_<SEEDLENGTH>-<READLENGTH>.txt.
Since the alignment program has been evolving, we have had different formats at different time points. Please refer to this page for the different formats handed out by GTC - http://jura.wi.mit.edu/genomecorewiki/index.php/AlignmentFormat
YLF (Young Lab Format) Files
These files are created on request only. The can be found in the "Eland_Results_Extended" folder along with the alignment results.